About
I am a Machine Learning Research Scientist at Bayer, where I work on machine learning applications to histopathology and drug discovery. Previously, I was a Postdoc at EMBL Heidelberg and a Visiting Scientist at Fraunhofer HHI. I received my PhD from Heidelberg University under the supervision of Anna Kreshuk and Fred Hamprecht. I did my MS in Computer Science at AGH University of Science and Technology, where I worked with Robert Schaefer. I create and contribute to popular open source machine learning projects. Sometimes I DJ.
Selected Publications
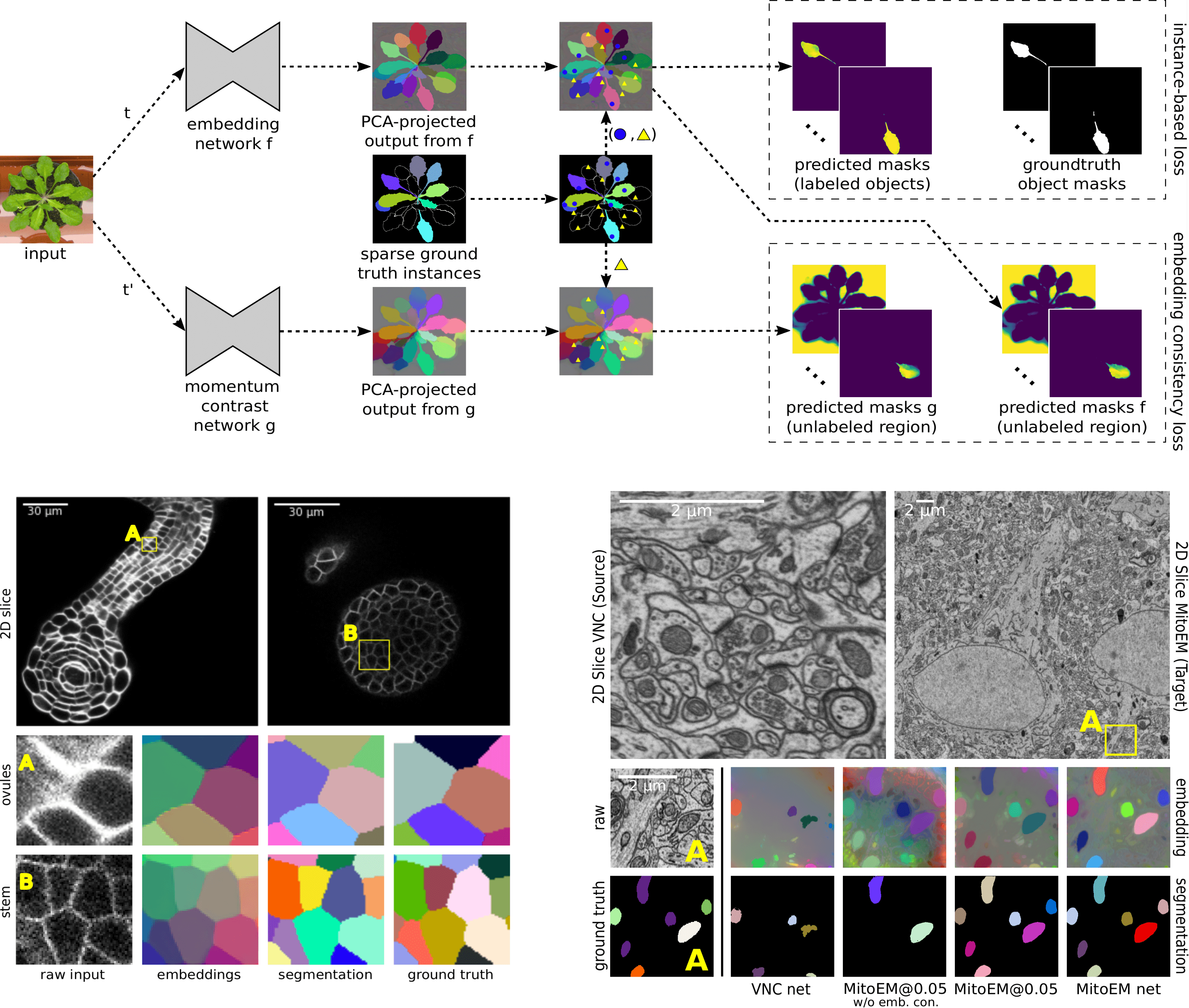
Most state-of-the-art instance segmentation methods have to be trained on densely annotated images. While difficult in general, this requirement is especially daunting for biomedical images, where domain expertise is often required for annotation and no large public data collections are available for pre-training. We propose to address the dense annotation bottleneck by introducing a proposal-free segmentation approach based on non-spatial embeddings, which exploits the structure of the learned embedding space to extract individual instances in a differentiable way. The segmentation loss can then be applied directly to instances and the overall pipeline can be trained in a fully- or weakly supervised manner. We consider the challenging case of positive-unlabeled supervision, where a novel self-supervised consistency loss is introduced for the unlabeled parts of the training data. We evaluate the proposed method on 2D and 3D segmentation problems in different microscopy modalities as well as on the Cityscapes and CVPPP instance segmentation benchmarks, achieving state-of-the-art results on the latter.
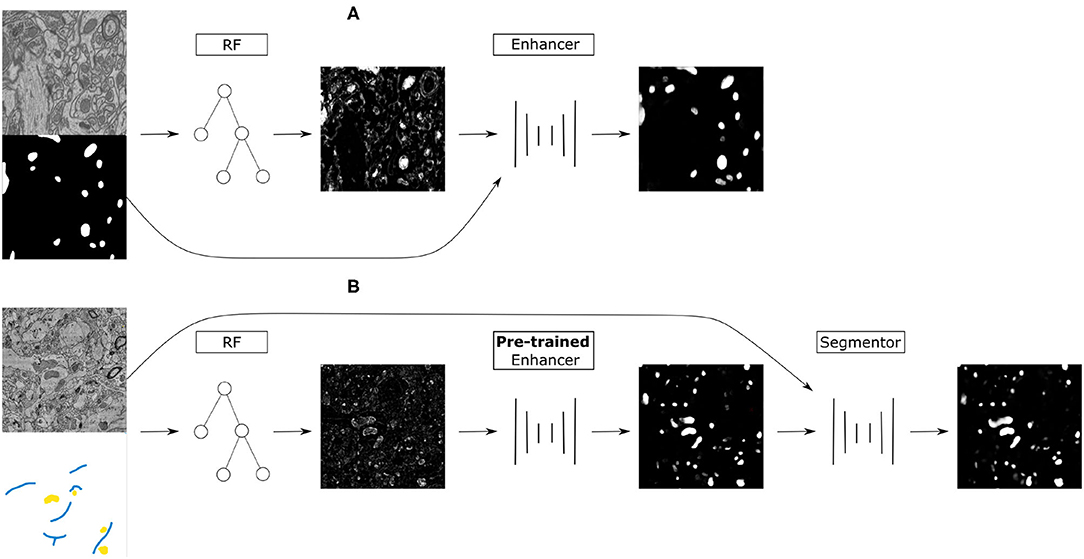
The remarkable performance of Convolutional Neural Networks on image segmentation tasks comes at the cost of a large amount of pixelwise annotated images that have to be segmented for training. In contrast, feature-based learning methods, such as the Random Forest, require little training data, but rarely reach the segmentation accuracy of CNNs. This work bridges the two approaches in a transfer learning setting. We show that a CNN can be trained to correct the errors of the Random Forest in the source domain and then be applied to correct such errors in the target domain without retraining, as the domain shift between the Random Forest predictions is much smaller than between the raw data. By leveraging a few brushstrokes as annotations in the target domain, the method can deliver segmentations that are sufficiently accurate to act as pseudo-labels for target-domain CNN training. We demonstrate the performance of the method on several datasets with the challenging tasks of mitochondria, membrane and nuclear segmentation. It yields excellent performance compared to microscopy domain adaptation baselines, especially when a significant domain shift is involved.
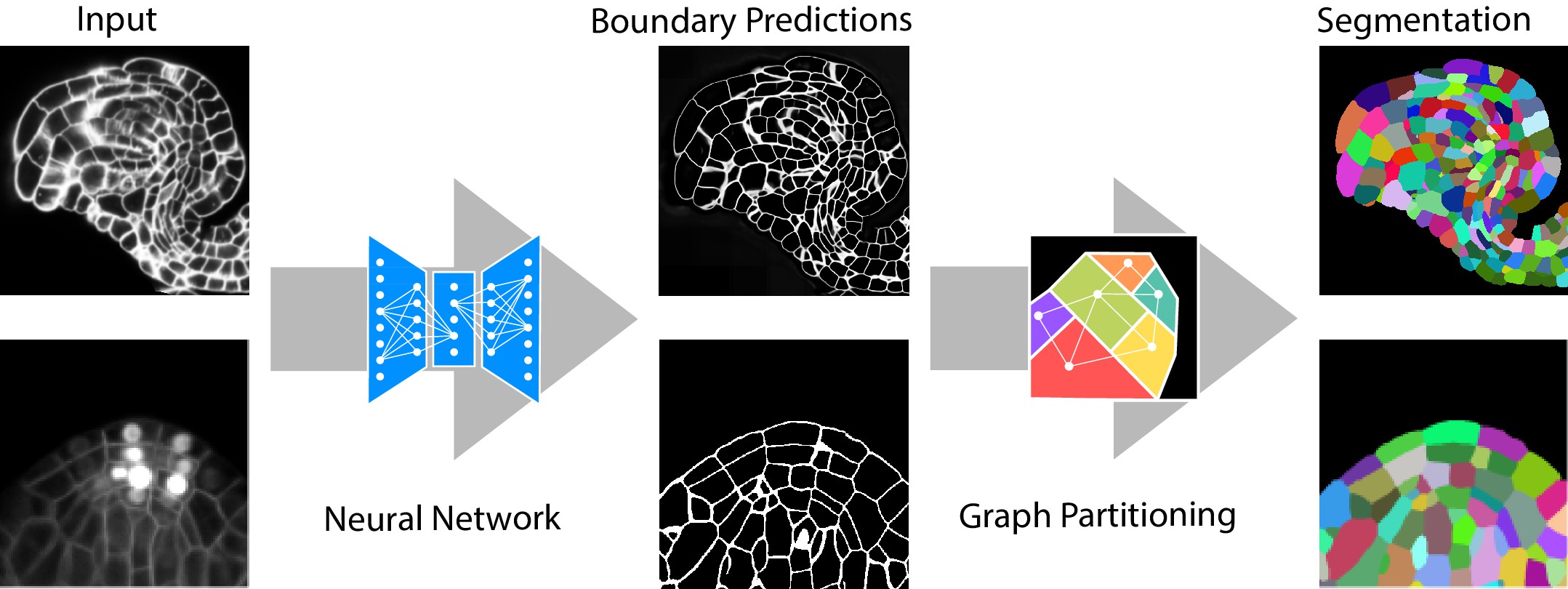
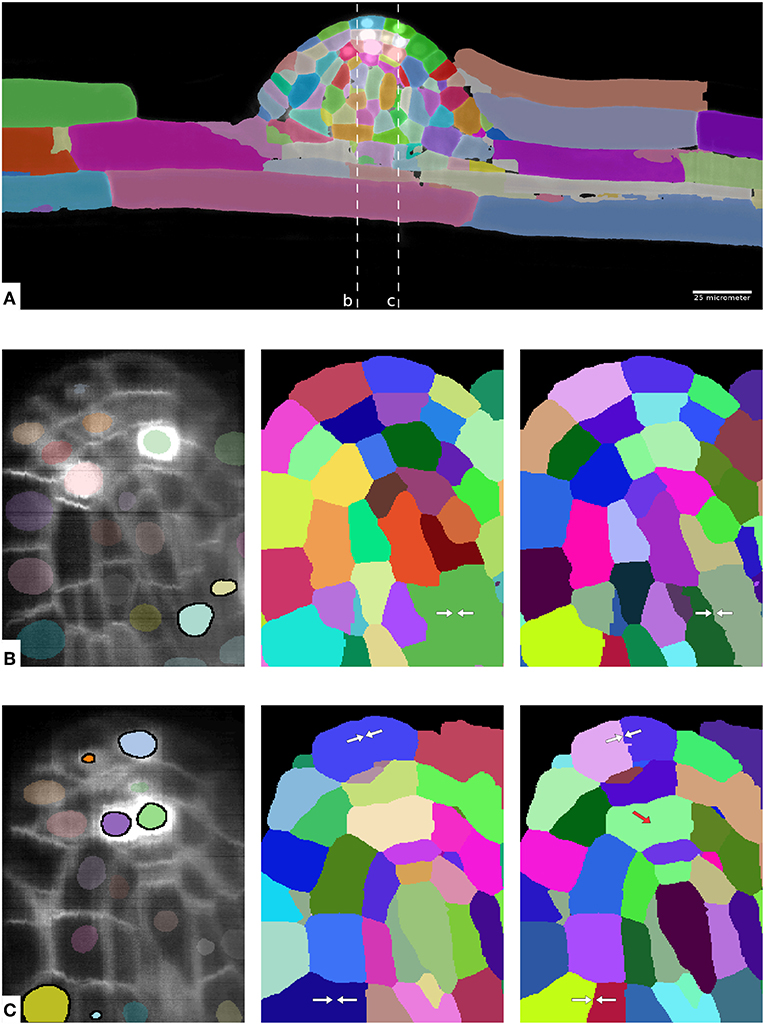
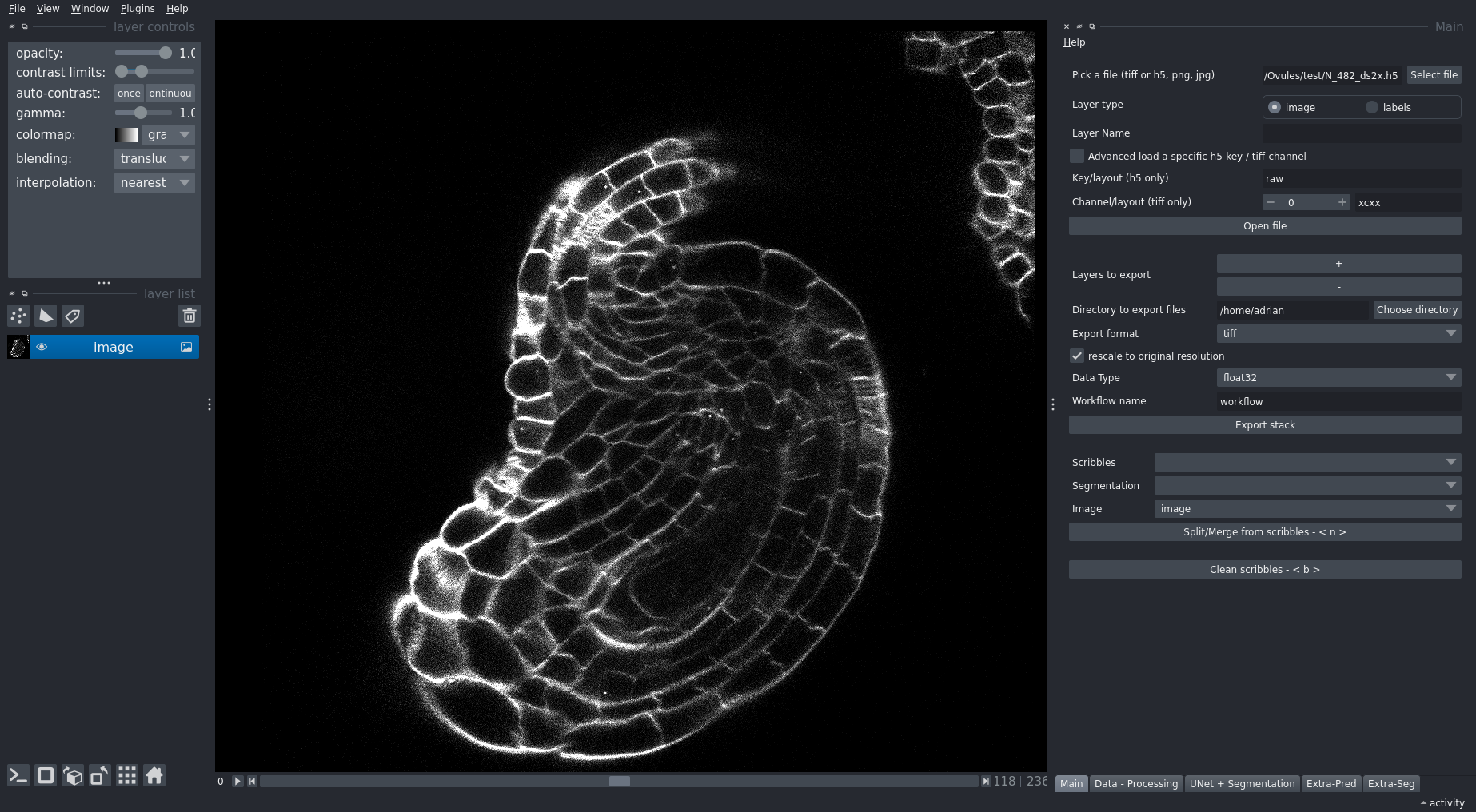
Quantitative analysis of plant and animal morphogenesis requires accurate segmentation of individual cells in volumetric images of growing organs. In the last years, deep learning has provided robust automated algorithms that approach human performance, with applications to bio-image analysis now starting to emerge. Here, we present PlantSeg, a pipeline for volumetric segmentation of plant tissues into cells. PlantSeg employs a convolutional neural network to predict cell boundaries and graph partitioning to segment cells based on the neural network predictions. PlantSeg was trained on fixed and live plant organs imaged with confocal and light sheet microscopes. PlantSeg delivers accurate results and generalizes well across different tissues, scales, acquisition settings even on non plant samples. We present results of PlantSeg applications in diverse developmental contexts. PlantSeg is free and open-source, with both a command line and a user-friendly graphical interface.
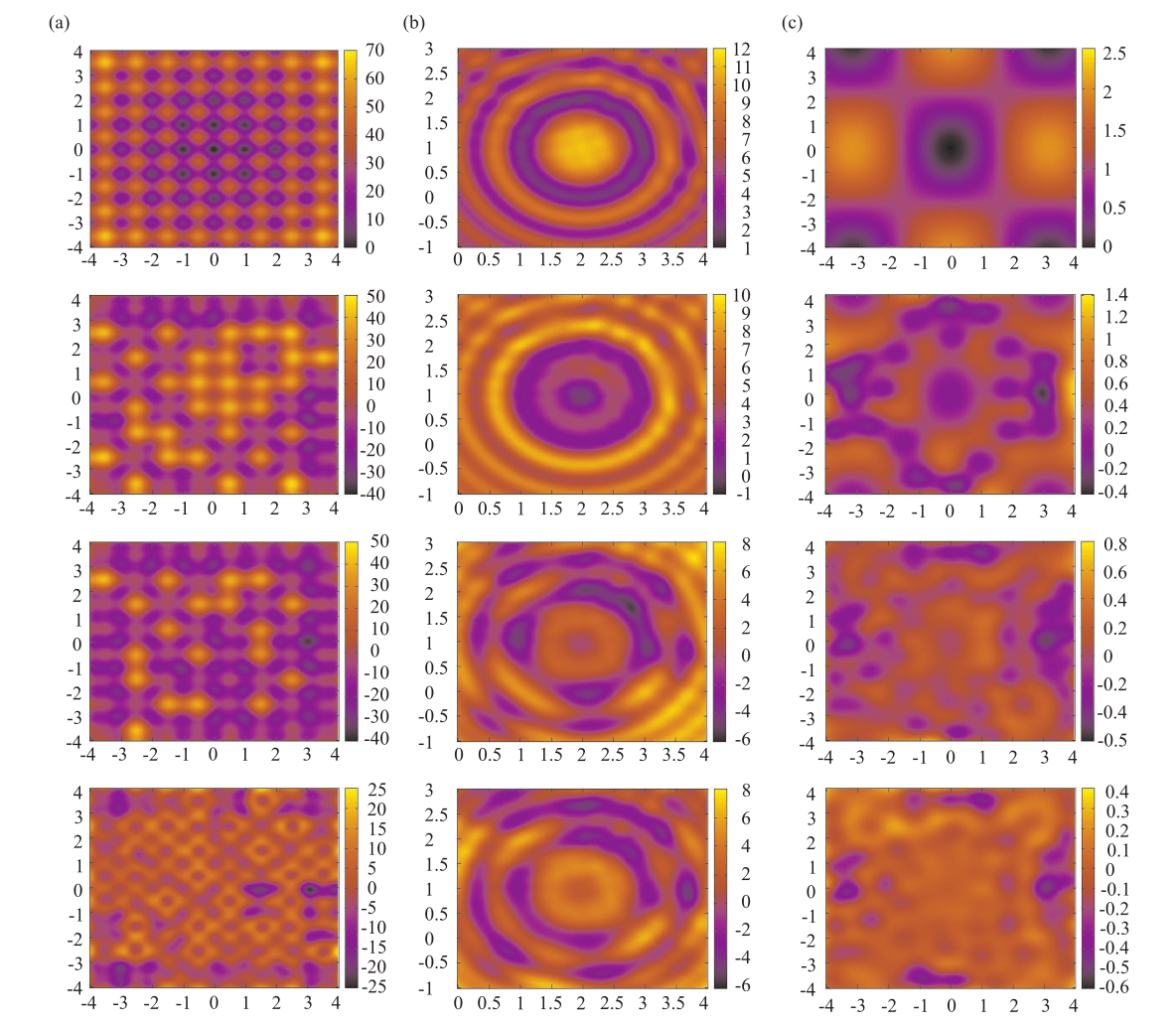
The throughput of electron microscopes has increased significantly in recent years, enabling detailed analysis of cell morphology and ultrastructure. Analysis of neural circuits at single-synapse resolution remains the flagship target of this technique, but applications to cell and developmental biology are also starting to emerge at scale. The amount of data acquired in such studies makes manual instance segmentation, a fundamental step in many analysis pipelines, impossible. While automatic segmentation approaches have improved significantly thanks to the adoption of convolutional neural networks, their accuracy still lags behind human annotations and requires additional manual proof-reading. A major hindrance to further improvements is the limited field of view of the segmentation networks preventing them from exploiting the expected cell morphology or other prior biological knowledge which humans use to inform their segmentation decisions. In this contribution, we show how such domain-specific information can be leveraged by expressing it as long-range interactions in a graph partitioning problem known as the lifted multicut problem. Using this formulation, we demonstrate significant improvement in segmentation accuracy for three challenging EM segmentation problems from neuroscience and cell biology.
This work presents a new hybrid approach for supporting sequential niching strategies called Cluster Supported Fitness Deterioration (CSFD). Sequential niching is one of the most promising evolutionary strategies for analyzing multimodal global optimization problems in the continuous domains embedded in the vector metric spaces. In each iteration CSFD performs the clustering of the random sample by OPTICS algorithm and then deteriorates the fitness on the area occupied by clusters. The selection pressure pushes away the next-step sample (population) from the basins of attraction of minimizers already recognized, speeding up finding the new ones. The main advantages of CSFD are low memory an computational complexity even in case of large dimensional problems and high accuracy of deterioration obtained by the flexible cluster definition delivered by OPTICS. The paper contains the broad discussion of niching strategies, detailed definition of CSFD and the series of the simple comparative tests.
Open Source Software
pytorch-3dunet is a popular Pytorch implementation of the standard 3D U-Net and its variant Residual 3D U-Net for volumetric semantic segmentation. It provides common functionalities for neural network training and inference accessible via a convenient config-based interface. It has been widely used by researchers and machine learning practitioners.
PlantSeg is a tool for cell segmentation in densely packed 3D images. It works best for images acquired with confocal and light-sheet microscopy. It is a two stage pipeline: CNN prediction followed by a graph partitioning. A large repository of pre-trained CNNs comes with the package.
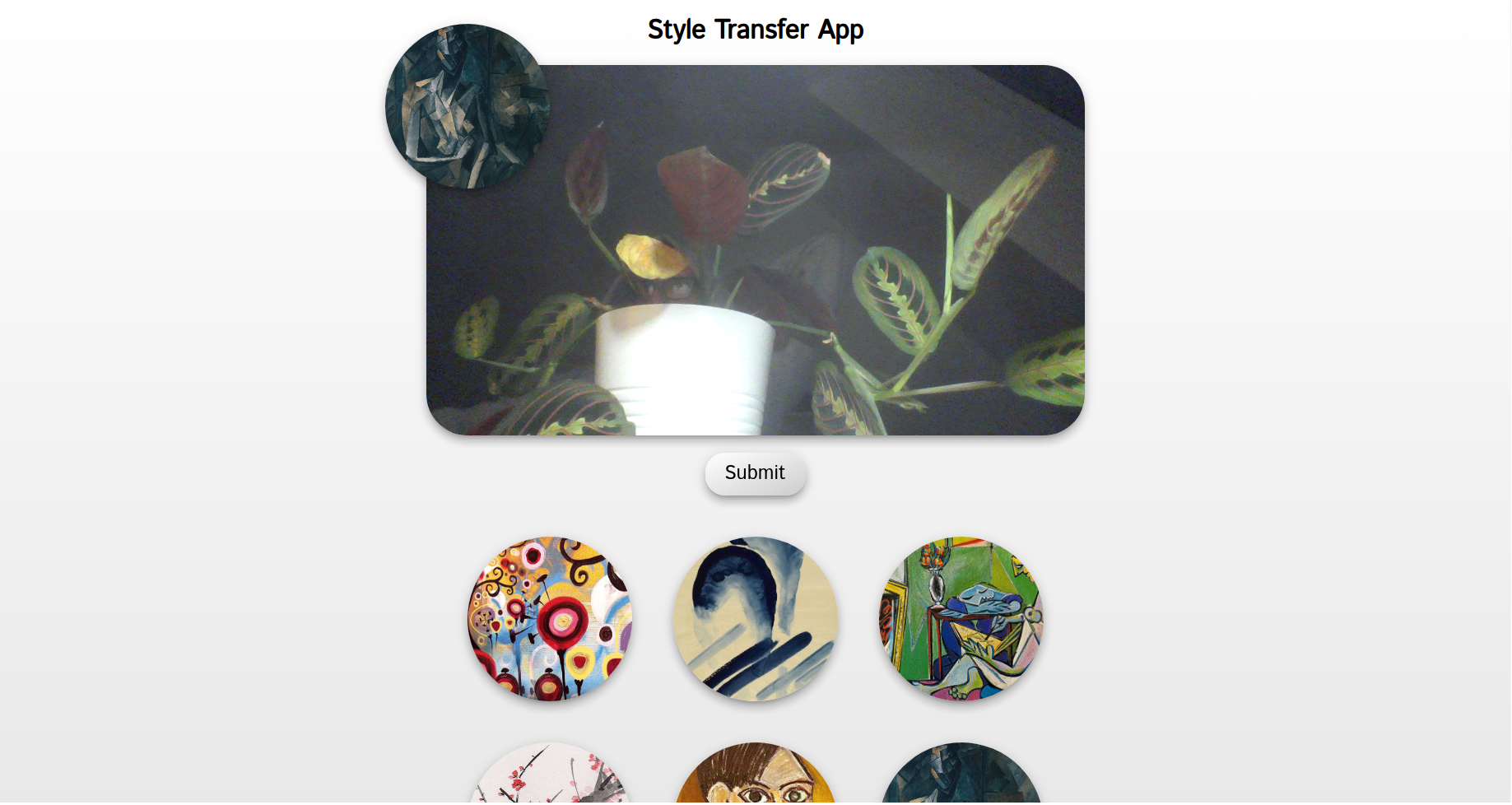
A simple web app which allows the user to take a photo with the webcam and apply a neural style transfer to it.